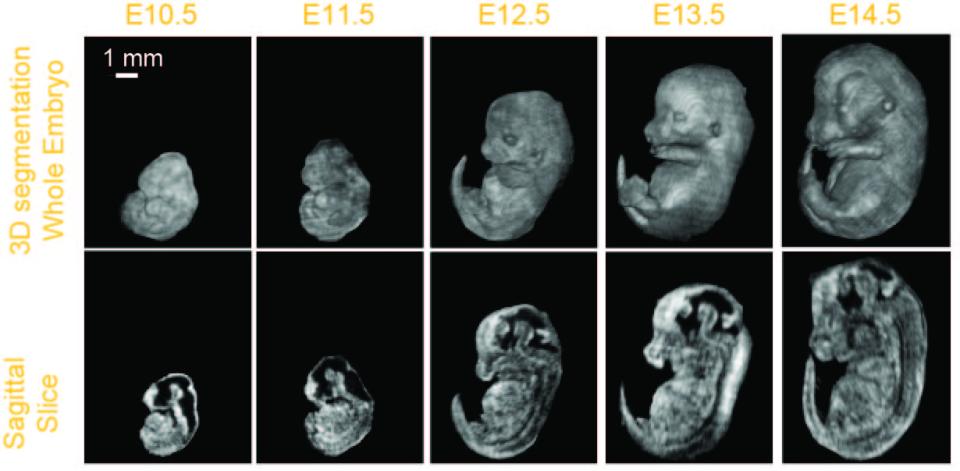
The goal of this proposal: phenotype early- to mid-gestational mouse embryos by segmenting select organ systems in 3D data sets acquired in utero with high-frequency ultrasound (HFU). The International Mouse Phenotyping Consortium (IMPC), which includes the NIH Knockout (KO) Mouse Phenotyping Program (KOMP2), will generate 20,000 mouse strains in the next decade, including many important models of human structural birth defects and congenital diseases. Developing phenotyping methods for efficient pipeline analyses of defects in embryonic growth in the KO mouse strains is a high priority for this effort. An in-utero 3D imaging approach, enabling volumetric and longitudinal analyses of various organ systems over a range of early- to mid-gestational stage mouse embryos, would provide added benefit and critical additional in vivo data not currently available.
Commercial HFU systems are widely available in many research centers largely thanks to the NIH-funded Small Animal Imaging Research Programs and Shared Instrumentation Programs. HFU is an excellent candidate modality to provide in utero 3D image data that can be quantitatively analyzed and archived to support the KOMP2/IMPC embryonic lethal phenotyping pipeline and future phenotyping efforts. We will develop and validate in utero 3D HFU image-acquisition protocols and image-processing methods that permit noninvasive, longitudinal studies of embryonic development and the detection and characterization of KO phenotypes. Volumetric HFU data will be collected in utero from mouse embryos staged between E9.5 to 15.5 to establish a database of normal development. Algorithms will be developed to segment 3D regions and extract parameters that quantify embryonic stage and identify regional changes between normal and KO embryos. We will acquire data with a custom, annular-array system and a VisualSonics Vevo 2100. The fine-resolution annular-array data will be used to initially develop the image-processing algorithms, and then the algorithms will be adapted for Vevo 2100 data. We will compare the quantitative parameters derived from the segmentation results obtained from the two scanners to ensure that the Vevo 2100 can provide equivalent mutant detection and quantification. Initial testing will be undertaken using wild-type and En1 and Gli2 mutants with known defects. Finally, the acquisition and processing protocols will be applied to 3D Vevo 2100 data from 5-10 KOMP2 KO mouse lines with embryonic defects in various organ systems to validate the HFU methods for detecting and characterizing phenotypes in these mutant embryos.