Award or grant: National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINNDS): R21 NS104634-01
Multiple Sclerosis (MS) damages white matter pathways that connect brain regions, i.e., the structural connectome (SC), disrupting flow of electrical signals, i.e., the functional connectome (FC), causing cognitive and physical disability. However, the burden of disease in the brain is not always proportional to an individual's disability. How the brain compensates for damage in resilient patients remains a mystery, making it difficult to develop accurate prognoses and treatments that can leverage this process in less fortunate patients. Without this knowledge, it will not be possible to create individualized therapies based on the brain's natural resiliency mechanisms, or to establish reliable ways to predict recovery potential. The thriving field of brain connectivity network analysis, or connectomics, provides a promising tool with which to capture, model and understand resiliency mechanisms. The lab’s long-term goal is to develop novel, personalized rehabilitation methods that mimic and enhance the brain's resiliency process to restore cognitive and physical abilities after damage due to neurological conditions. The objective of this work (being conducted with the team of Susan Gauthier, D.O.): to identify MS connectome-based imaging biomarkers of resiliency, i.e., those separating patients with high disease burden and low disability (high-adapters) from those with similar disease burden and high disability (low-adapters). The rationale for the proposed research: insight into the brain's ability to compensate for injury will lead to more accurate prognostication and enable development of novel therapeutic MS strategies.
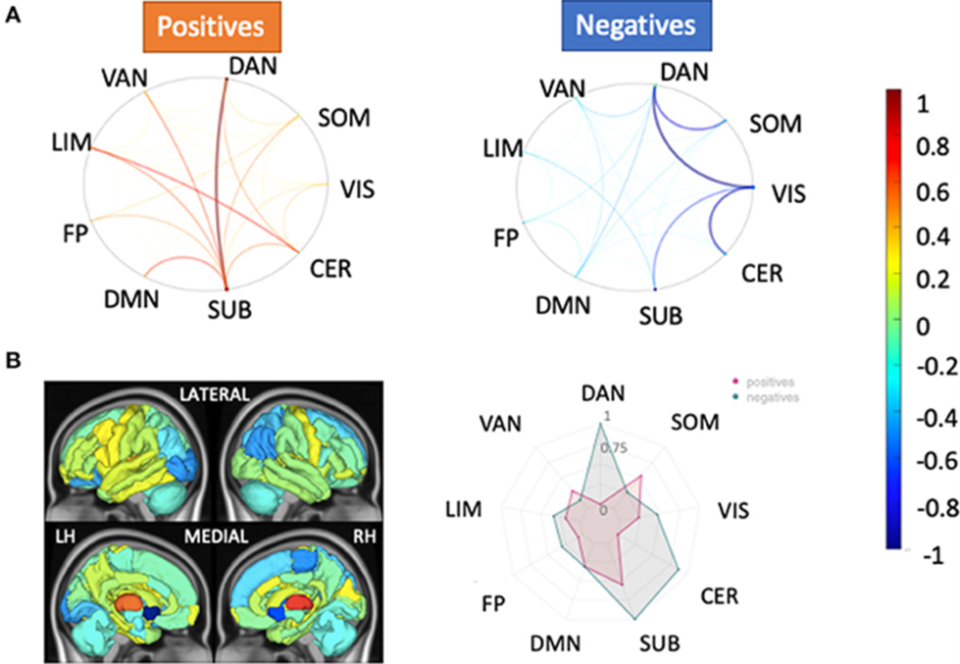
The lab pursued two Specific Aims: to identify global and regional metrics of the 1) structural and functional connectomes and 2) structure-function relationship between the connectomes that differentiate high-adapting and low-adapting MS patients. The grant led to two peer-reviewed journal publications, a third manuscript in preparation, and oral/poster presentations at many conferences. The lab collected functional, diffusion, and anatomical magnetic resonance images (MRIs) from 20 controls and 100 people with MS (pwMS). The lab processed their brain images, extracted various metrics of SC and static and dynamic FC, and compared these metrics between controls and pwMS and pwMS with and without disability. The lab found SC was the metric most different between controls vs. MS, while dynamic FC was most different between pwMS with and without disability. This hints that, while anatomical damage occurs across the MS population compared to controls, individual MS patients’ brains may compensate by reorganizing their functional brain activity patterns. Tozlu C., et al., Dynamic functional connectivity better predicts disability than structural and static functional connectivity in people with multiple sclerosis, Front. Neurosci., 15:763966: 2021. (Please see the above image, which depicts relative feature weights of the structural connectivity (SC) models for healthy controls (HC) vs. people with multiple sclerosis (pwMS) classification task. Relative feature weights (scaled by the maximum magnitude feature weight) for the variables used in the two models with the best classification performance in the HC vs. pwMS task: (A) pairwise SC and (B) regional SC (node strength). The circle plots in (A) illustrate the positive (hot colors) and negative (cool colors) model feature weights, respectively, for the pairwise SC model, averaged across the Yeo functional networks. The glass brain and radial plot figures in (B) show the relative feature weights from the regional SC (node strength) model, where the redial plot shows the positive and negative values averaged over the Yeo functional networks. Negative values (cooler colors) show those connections where larger values were associated with greater probability of being in the HC group, while positive values (hotter colors) show those connections where larger values were associated with greater probability of being in the pwMS group.) The lab also found:
- PwMS spent increased time in brain activity states with high somatomotor activation compared to controls, possibly indicating functional upregulation or compensatory activity in motor-related regions, while the functional activity in those regions decreases in patients with greater disability (manuscript in preparation).
- Machine learning models based on estimates of the SC and FC metrics better-classified pwMS by disability level than SC and FC observed directly in the individual using advanced magnetic resonance imaging (MRI). This demonstrates a clinically feasible alternative to high-cost, advanced MRI in patient populations that could be used to better understand brain lesion-dysfunction mapping and make more accurate prognoses. Tozlu, C., et al., Estimated connectivity networks outperform observed connectivity networks when classifying people with multiple sclerosis into high and low disability groups, NeuroImage: Clinical. (32): 102827: 2021.
The quantitative methods applied here to multi-modal connectome imaging in a large cohort of pwMS helped in understanding of disease and compensatory mechanisms, enabling more accurate prognoses, and possibly the future development of novel therapeutics to promote activity in somato-motor regions in pwMS and promote recovery.